SurfStamp: 3D Printer Compatible Surface Representation of Macromolecules
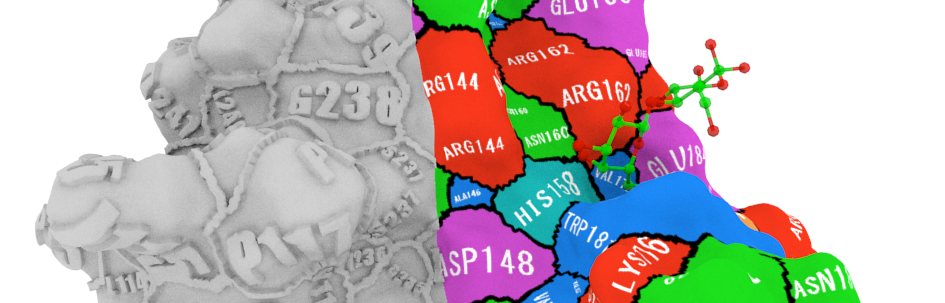
SurfStamp is an application to generate textures for surface models of proteins. The information of residues; residue types and numbers are drawn directly on the 3D objects. This approach is more intuitive than the labeling functions that most of 3D structure viewers use to show residue information.
Thus, with this application, researchers, readers or audiences can easily distinguish which residues are contributing the surface they are focusing on.
This application is distributed under Apache License Version 2.0. You can freely use, copy, modify and distribute as long as you follow the license terms.
Thus, with this application, researchers, readers or audiences can easily distinguish which residues are contributing the surface they are focusing on.
This application is distributed under Apache License Version 2.0. You can freely use, copy, modify and distribute as long as you follow the license terms.
Download:
Check the latest releaseUsage:
SurfStamp has function to generate surface model using procedure like EDTSurf (but a bit different, the difference may be introduced in future).
In addition, you can also use models made by other software.
But please be careful because some GUI type software generate objects according to their camera position.
See tips↓ for detail.
SurfStamp is coded 100% Java language. You can run it on any OS as long as JavaVM is installed.
Example command is as follows.
In addition, you can also use models made by other software.
But please be careful because some GUI type software generate objects according to their camera position.
See tips↓ for detail.
SurfStamp is coded 100% Java language. You can run it on any OS as long as JavaVM is installed.
Example command is as follows.
java -jar SurfStamp.jar -pdb 3zsj_prot.pdb -nowater -ligand 3zsj_ligand.pdb -out 3zsj_example.obj -color_scheme 0 -image_size 4096
3zsj_prot.pdb: ATOM record of the protein (Galectin-3 CRD) in 3ZSJ.pdb.
3zsj_ligand.pdb: HETATM record of the BETA-LACTOSE in 3ZSJ.pdb.
Please download SurfStamp_with_example.zip from release page. The files are in it.
3zsj_ligand.pdb: HETATM record of the BETA-LACTOSE in 3ZSJ.pdb.
Please download SurfStamp_with_example.zip from release page. The files are in it.
OutputExample:
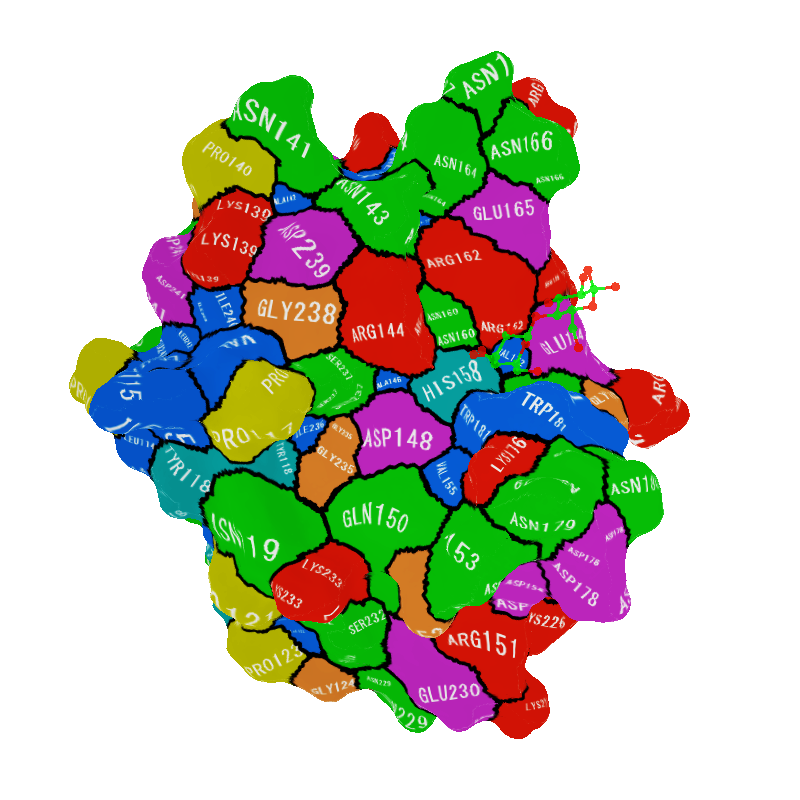
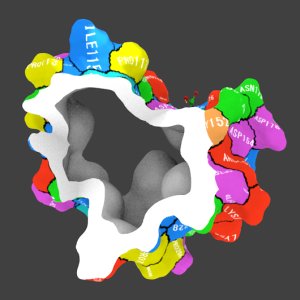
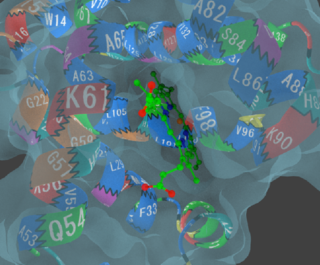
3D object of 3ZSJ made with the above command.
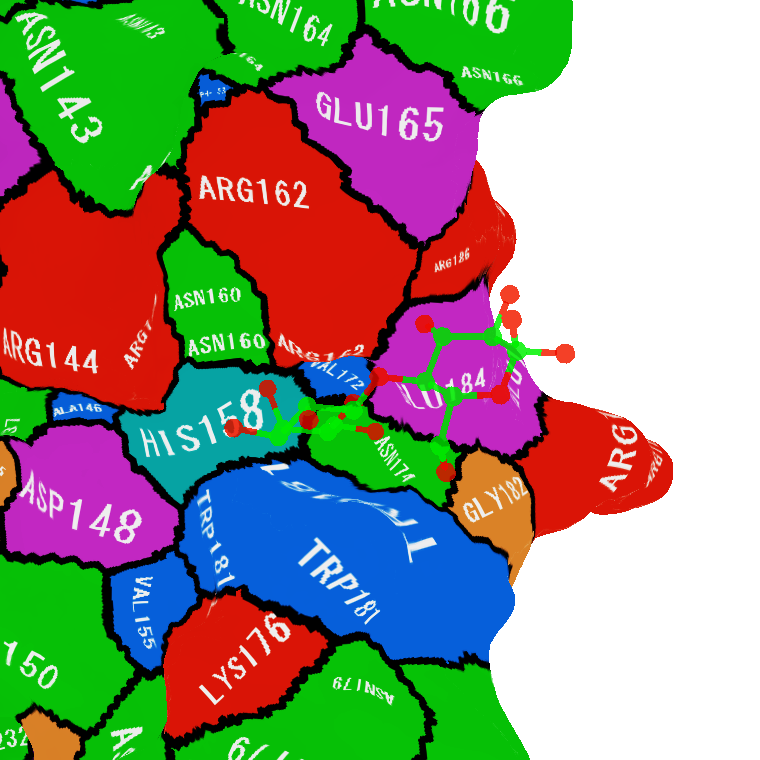
You can see residues interacting with ligands at a glance.
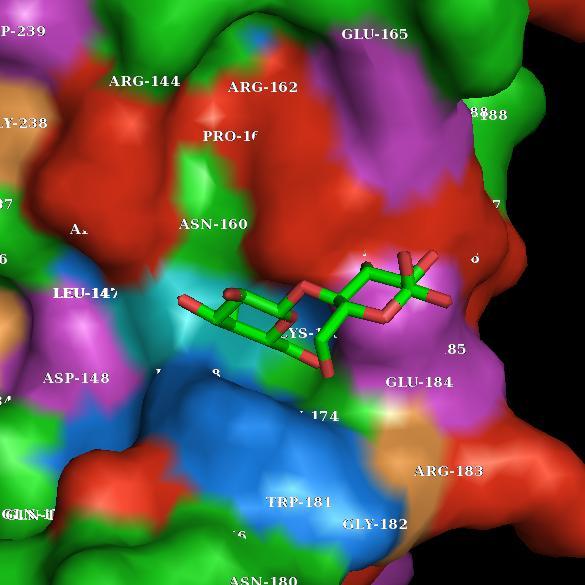
I did my best to make the same object using PyMol.

Another example: 3D object of 3DF6 (chain A).
There is a Threonine cluster... Is it phospholylated?
SurfStamp will provide clues for further experiments like this.

Another example2: CPK coloring (-color_scheme 3) of 3DF6 (chain A).
More images
 |
 |
 |
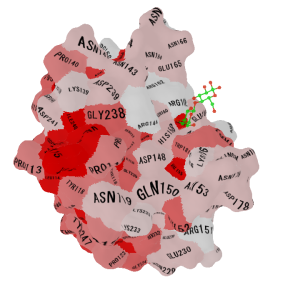
| -color_scheme 1 & -nooutline (Hydrophobisity, Red: hydrophobic) |
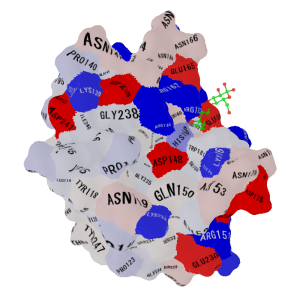
-color_scheme 2 & -nooutline (Isoelectric point, Red: acidic, Blue: basic) |
-lighteningwall (Lightening to reduce the cost of 3D print) |
 |
 |
 |
| The first 3d printed model. x2.5 using DMM 3d print service. The printing cost was appox. $400. |
Since DMM 3D print service does not consider inner space of the object, I divided the model into 2 parts with blender. | The resolution rate is not so high... I think I should have made model with "-residue_oneletter". |
| The model I printed can be downloaded from NIH 3DPrint Exchange. http://3dprint.nih.gov/discover/3dpx-003891 (Click download icon and download zip file(s).) | ||
 |
 |
 |
| -font_name Impact -residue_oneletter -text_weight plain -extrude 0.35 -lighteningwall Residue information can be extruded or intruded for mono color 3d print. |
Approximately $140 with Nylon using DMM 3d print service. | The resolution rate was enough to see the residue names but it may be good to paint important residues manually. |
| The model I printed can be downloaded from NIH 3DPrint Exchange. http://3dprint.nih.gov/discover/3dpx-004144 (Click download icon and download zip file(s).) | ||
 |
 |
 |
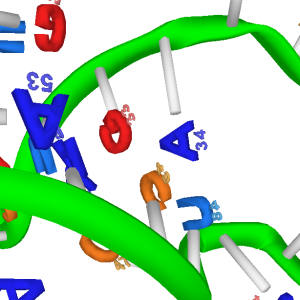
| Fancy polynucleotide model is available from v0.10 with -dnarna option. The letters have almost same size with that of six-membered ring (Oracle java 1.8.0_121 in Windows 8.1). | Watson-Crick pairs are intuitively seen by users. | Cryo-EM structure of the spliceosome 5lj3. (large image) Surfstamp approach is even more useful to understand archtecture of large macromolecule complex. |
 |
 |
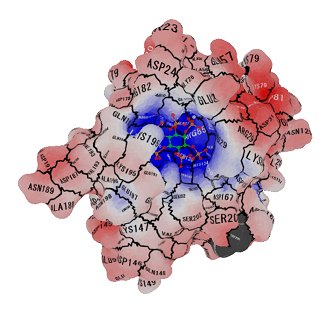
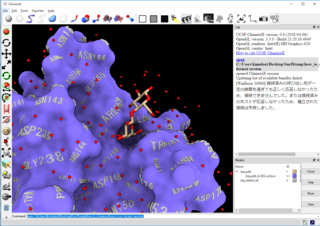 |
| Result of APBS (.dx file) can be imported with -color_dx option. | Of course it can be printed out by 3D printer! NIH 3D Print Exchange | You can make ChimeraX compatible output with -text_object option! |
 |
||
| Combined with Kawakami model style. (Cartoon model by Pymol & -residue_oneletter -usebackbone options) | ||
Check other structures (not github site & not https):
Tips:
Making 3D geometry (.obj file) with PyMol(CUI).
If you can use PyMol from terminal.
ref: https://www.biostars.org/p/42474/
https://pymolwiki.org/index.php/Launching_From_a_Script
1. Save following script as "objmake.py".
3. XXXX.obj will be saved.
ref: https://www.biostars.org/p/42474/
https://pymolwiki.org/index.php/Launching_From_a_Script
1. Save following script as "objmake.py".
import __main__2. Call "python objmake.py XXXX.pdb".
__main__.pymol_argv = [ 'pymol', '-qc']
import sys, time, os
import pymol
from chempy import models, cpv
pymol.finish_launching()
spath = sys.argv[1]
sname = spath.split('/')[-1].split('.')[0]
# Load Structures
print(spath);
pymol.cmd.load(spath,sname);
pymol.cmd.hide("all");
pymol.cmd.show("surface");
pymol.cmd.reset();
pymol.cmd.origin(position=[0.0,0.0,0.0]);
# pymol.cmd.center(origin = 0);
pymol.cmd.pseudoatom("pseudo_",pos=[0,0,0]);
pymol.cmd.select("psel","/pseudo_//P/PSD`1/PS1");
pymol.cmd.center(selection="psel");
pymol.cmd.save(sname+".obj");
pymol.cmd.delete("all");
pymol.cmd.quit()
3. XXXX.obj will be saved.
Making 3D geometry (.obj file) with PyMol(GUI).
1. Extract only ATOM and HETATM lines (with grep or manually) and save them as XXXX_atomlines.pdb or any names you want.
2. Load XXXX_atomlines.pdb.
2-b. Do not perform any transform operation (rotate, translate, expand and so forth).
3. Make surface model with [S]->surface.
4. Input following command.
2. Load XXXX_atomlines.pdb.
2-b. Do not perform any transform operation (rotate, translate, expand and so forth).
3. Make surface model with [S]->surface.
4. Input following command.
origin position=[0,0,0]5. XXXX.obj will be saved.
center origin
save XXXX.obj
Make 3D geometry (.ply file) with EDTSurf.
Please follow the instruction of EDTSurf.
Make 3D geometry (.obj file) with UCSF Chimera.
1. Load XXXX.pdb.
2. Make surface model with [Actions]->[Surface]->[Show].
2-b. or enter "surf #0 grid 0.5" in the command line. Where 0 is the model number and 0.5 is the mesh size.
3. Hide all other models with [Actions]->[Atoms/Bonds]->[Hide] and [Actions]->[Ribbon]->[Hide].
4. If you have rotated or moved the model, enter "reset" in the command line ([Tools]->[General controls]->[Command line])).
(As I do not use UCSF Chimera frequently, the operation may not be enough. Please let me know if you can't make the model correctly.)
5. Save the model with [File]->[Export Scene]-> choose "OBJ [.obj]" in the File type combobox -> enter filename and press [Save].
2. Make surface model with [Actions]->[Surface]->[Show].
2-b. or enter "surf #0 grid 0.5" in the command line. Where 0 is the model number and 0.5 is the mesh size.
3. Hide all other models with [Actions]->[Atoms/Bonds]->[Hide] and [Actions]->[Ribbon]->[Hide].
4. If you have rotated or moved the model, enter "reset" in the command line ([Tools]->[General controls]->[Command line])).
(As I do not use UCSF Chimera frequently, the operation may not be enough. Please let me know if you can't make the model correctly.)
5. Save the model with [File]->[Export Scene]-> choose "OBJ [.obj]" in the File type combobox -> enter filename and press [Save].
How to see the model with PyMOL.
Please try PyMOL plugin! https://github.com/yamule/SurfStamp-pymol-pluginLicense:
Copyright 2016-2020 yamule
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
I thank James Tyrwhitt Drake and other staff of NIH 3D Print Exchange for encouraging this work and helpful suggestions.
Licensed under the Apache License, Version 2.0 (the "License");
you may not use this file except in compliance with the License.
You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software
distributed under the License is distributed on an "AS IS" BASIS,
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
See the License for the specific language governing permissions and
limitations under the License.
I thank James Tyrwhitt Drake and other staff of NIH 3D Print Exchange for encouraging this work and helpful suggestions.